This vignette shows the basics of using watlastools. It is a work in progress. watlastools has two main purposes: accessing and cleaning data, and identifying residence patches from the data. Only the latter is covered here.
library(watlastools)
Making residence patches
Residence patches are the end goal of the watlastools workflow. They are a conveninent way of synthesising large amounts of tracking data into a few spatially explicit units that have a set of associated (mean) coordinates, as well as ecologically relevant coordinates such as duration.
Begin by reading in the data.
# read data and see leading lines library(data.table) data <- fread("vignette_data/413_004_revisit.csv") head(data) #> x y coordIdx temp_time ts #> 1: 646876.3 5903885 1 2018-08-12T13:30:00Z 2018-08-12T13:25:29.634667Z #> 2: 646810.2 5904035 2 2018-08-12T13:30:00Z 2018-08-12T13:25:46.634Z #> 3: 646929.2 5903835 3 2018-08-12T13:30:00Z 2018-08-12T13:32:37.615Z #> 4: 646877.9 5903939 4 2018-08-12T13:30:00Z 2018-08-12T13:32:57.114500Z #> 5: 646930.5 5903887 5 2018-08-12T13:40:00Z 2018-08-12T13:36:25.605Z #> 6: 646972.9 5903816 6 2018-08-12T13:40:00Z 2018-08-12T13:36:57.603333Z #> time id posID X_raw Y_raw NBS VARX VARY COVXY #> 1: 1534080330 413 3.0 646876.3 5903885 3 1409.000 6058.883 -2880.460 #> 2: 1534080360 413 5.0 646810.2 5904035 3 2338.890 9407.620 -4618.900 #> 3: 1534080750 413 8.0 646929.2 5903835 3 609.179 2814.890 -1295.480 #> 4: 1534080780 413 9.5 646877.9 5903939 3 1136.403 4910.655 -2330.560 #> 5: 1534080990 413 11.0 646930.5 5903887 3 1402.250 6461.530 -2989.980 #> 6: 1534081020 413 13.0 646972.9 5903816 3 1086.319 5678.337 -2437.807 #> SD tide_number tidaltime waterlevel resTime fpt revisits #> 1: 41.21546 4 335.4939 -106 0.08615906 0.08615906 1 #> 2: 50.08702 4 335.7772 -106 1.55416984 1.55416984 1 #> 3: 28.86363 4 342.6269 -106 5.72246712 1.60784093 2 #> 4: 36.92465 4 342.9519 -106 2.49882594 2.49882594 1 #> 5: 43.40300 4 346.4268 -113 2.68004833 2.68004833 1 #> 6: 43.25045 4 346.9601 -113 7.87090615 0.09736257 2
Now make residence patches after inferring residence from gaps in the data.
# run the three main functions on the data # first infer residence temp_data <- wat_infer_residence(data = data, inf_patch_time_diff = 30, inf_patch_spat_diff = 100) # classify residence points temp_data <- wat_classify_points(data = temp_data, lim_res_time = 2) # make residence patches with sensible parameters patches <- wat_make_res_patch(data = temp_data, buffer_radius = 10, lim_spat_indep = 100, lim_time_indep = 30, lim_rest_indep = 30, min_fixes = 3)
Getting data from patches
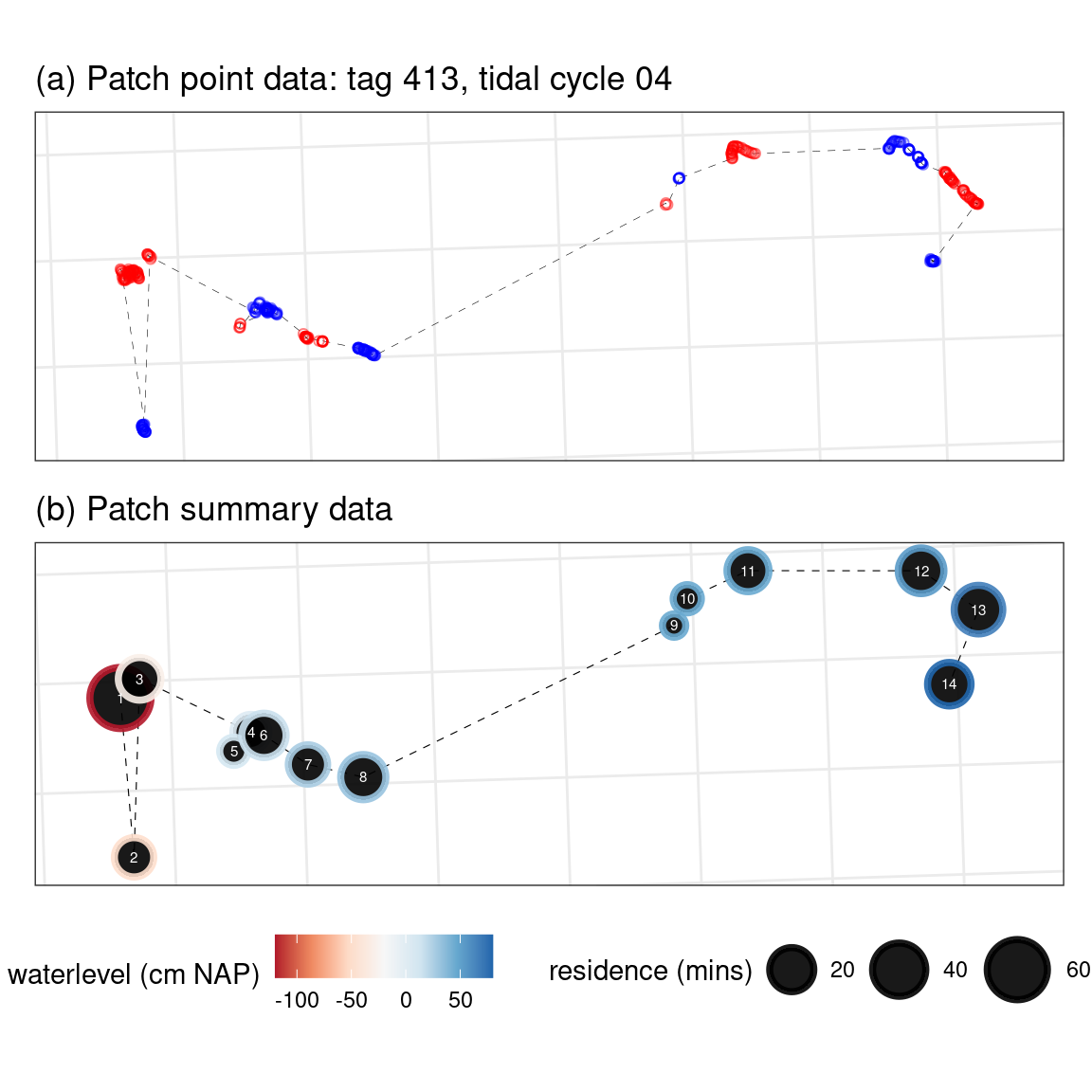
Get and view the different kinds of patch data
Patch summary
The patch summary is a data.table and has no complex (list, sf) column types. The patches can be visualised using ggplot (or anything else).
# get patch summary patch_summary <- wat_get_patch_summary(res_patch_data = patches, which_data = "summary") # get the class of the full object and the column types class(patch_summary) #> [1] "data.table" "data.frame"
Patch points
The patch points are the raw points that make up the residence patches.
# get patch points patch_points <- wat_get_patch_summary(res_patch_data = patches, which_data = "points") # get class and column type class(patch_points) #> [1] "data.table" "data.frame"
Patch polygons
This should have the added class sf, and the polygons column should have the type sfc_MULTIPOLYGON.
# get patch points patch_polygons <- wat_get_patch_summary(res_patch_data = patches, which_data = "spatial") # get class and column type class(patch_polygons) #> [1] "sf" "data.table" "data.frame"