Section 4 Working with lists and iteration

4.1 List columns with tidyr
4.1.1 Nesting data
It may become necessary to indicate the groups of a tibble in a somewhat more explicit way than simply using dplyr::group_by. tidyr offers the option to create nested tibbles, that is, to store complex objects in the columns of a tibble. This includes other tibbles, as well as model objects and plots.
NB: Nesting data is done using tidyr::nest, which is different from the similarly named tidyr::nesting.
The example below shows how Phylacine data can be converted into a nested tibble.
# get phylacine data
data = read_csv("data/phylacine_traits.csv")
data = data %>%
`colnames<-`(str_to_lower(colnames(.))) %>%
`colnames<-`(str_remove(colnames(.), "(.1.2)")) %>%
`colnames<-`(str_replace_all(colnames(.), "\\.", "_"))# nest phylacine by order
nested_data = data %>%
group_by(order) %>%
nest()
nested_data
#> # A tibble: 29 x 2
#> # Groups: order [29]
#> order data
#> <chr> <list>
#> 1 Rodentia <tibble [2,306 × 23]>
#> 2 Chiroptera <tibble [1,162 × 23]>
#> 3 Carnivora <tibble [313 × 23]>
#> 4 Pilosa <tibble [34 × 23]>
#> 5 Diprotodontia <tibble [183 × 23]>
#> 6 Cetartiodactyla <tibble [392 × 23]>
#> # … with 23 more rows
# get column class
sapply(nested_data, class)
#> order data
#> "character" "list"The data is now a nested data frame. The class of each of its columns is respectively, a character (order name) and a list (the data of all mammals in the corresponding order).
While nest can be used without first grouping the tibble, it’s just much easier to group first.
4.1.2 Unnesting data
A nested tibble can be converted back into the original, or into a processed form, using tidyr::unnest. The original groups are retained.
# use unnest to recover the original data frame
unnest(nested_data, cols = "data") %>%
head()
#> # A tibble: 6 x 24
#> # Groups: order [1]
#> order binomial family genus species terrestrial marine freshwater aerial
#> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Rode… Abditom… Murid… Abdi… latide… 1 0 0 0
#> 2 Rode… Abeomel… Murid… Abeo… sevia 1 0 0 0
#> 3 Rode… Abraway… Crice… Abra… ruschii 1 0 0 0
#> 4 Rode… Abrocom… Abroc… Abro… bennet… 1 0 0 0
#> 5 Rode… Abrocom… Abroc… Abro… bolivi… 1 0 0 0
#> 6 Rode… Abrocom… Abroc… Abro… budini 1 0 0 0
#> # … with 15 more variables: life_habit_method <chr>, life_habit_source <chr>,
#> # mass_g <dbl>, mass_method <chr>, mass_source <chr>, mass_comparison <chr>,
#> # mass_comparison_source <chr>, island_endemicity <chr>, iucn_status <chr>,
#> # added_iucn_status <chr>, diet_plant <dbl>, diet_vertebrate <dbl>,
#> # diet_invertebrate <dbl>, diet_method <chr>, diet_source <chr>
# unnesting preserves groups
groups(unnest(nested_data, cols = "data"))
#> [[1]]
#> orderThe unnest_longer and unnest_wider variants of unnest are maturing functions, that is, not in their final form. They allow interesting variations on unnesting — these are shown here but advised against.
Unnest the data first, and then convert it to the form needed.
4.1.3 Working with list columns
The class of a list column is list, and working with list columns (and lists, and list-like objects such as vectors) makes iteration necessary, since this is one of the only ways to operate on lists.
Two examples are shown below when getting the class and number of rows of the nested tibbles in the list column.
# how many rows in each nested tibble?
for (i in seq_along(nested_data$data[1:10])) {
print(nrow(nested_data$data[[i]]))
}
#> [1] 2306
#> [1] 1162
#> [1] 313
#> [1] 34
#> [1] 183
#> [1] 392
#> [1] 460
#> [1] 57
#> [1] 20
#> [1] 465
# what is the class of each element?
lapply(X = nested_data$data[1:3], FUN = class)
#> [[1]]
#> [1] "tbl_df" "tbl" "data.frame"
#>
#> [[2]]
#> [1] "tbl_df" "tbl" "data.frame"
#>
#> [[3]]
#> [1] "tbl_df" "tbl" "data.frame"Functionals
The second example uses lapply, and this is a functional. Functionals are functions that take another function as one of their arguments. Base R functionals include the *apply family of functions: apply, lapply, vapply and so on.
4.2 Iteration with map
The tidyverse replaces traditional loop-based iteration with functionals from the purrr package.
Why use purrr
A good reason to use purrr functionals instead of base R functionals is their consistent and clear naming, which always indicates how they should be used. This is explained in the examples below. How map is different from for and lapply are best explained in the Advanced R Book.
4.2.1 Basic use of map
map works very similarly to lapply, where .x is object on whose elements to apply the function .f.
# get the number of rows in data
map(.x = nested_data$data, .f = nrow) %>%
head()
#> [[1]]
#> [1] 2306
#>
#> [[2]]
#> [1] 1162
#>
#> [[3]]
#> [1] 313
#>
#> [[4]]
#> [1] 34
#>
#> [[5]]
#> [1] 183
#>
#> [[6]]
#> [1] 392map works on any list-like object, which includes vectors, and always returns a list. map takes two arguments, the object on which to operate, and the function to apply to each element.
4.2.2 map variants returning vectors
Though map always returns a list, it has variants named map_* where the suffix indicates the return type.
map_chr, map_dbl, map_int, and map_lgl return character, double (numeric), integer, and logical vectors.
# use map_dbl to get the mean mass in each order
map_dbl(nested_data$data, function(df){
mean(df$mass_g)
})
#> [1] 4.86e+02 4.91e+01 4.79e+04 7.86e+05 4.02e+04 1.85e+06 6.68e+03 3.06e+02
#> [9] 1.61e+02 4.06e+01 7.48e+02 1.45e+03 2.36e+05 3.37e+01 1.74e+02 9.58e+05
#> [17] 9.03e+02 4.70e+06 1.13e+03 2.84e+03 2.23e+01 1.12e+06 1.83e+02 5.94e+05
#> [25] 1.22e+04 9.44e+03 1.65e+06 4.45e+01 5.24e+04
# map_chr will convert the output to a character
# here we get the most common IUCN status of each order
map_chr(nested_data$data, function(df){
count(df, iucn_status) %>%
arrange(-n) %>%
summarise(common_status = first(iucn_status)) %>%
pull(common_status)
})
#> [1] "LC" "LC" "LC" "EP" "LC" "LC" "LC" "LC" "LC" "LC" "LC" "LC" "EP" "VU" "LC"
#> [16] "EP" "LC" "EP" "LC" "LC" "NT" "VU" "LC" "EP" "VU" "CR" "EP" "LC" "LC"
# map_lgl returns TRUE/FALSE values
some_numbers = c(NA, 1:3, NA, NaN, Inf, -Inf)
map_lgl(some_numbers, is.na)
#> [1] TRUE FALSE FALSE FALSE TRUE TRUE FALSE FALSE4.2.3 map variants returning data frames
map_df returns data frames, and by default binds dataframes by rows, while map_dfr does this explicitly, and map_dfc does returns a dataframe bound by column.
# get the first two rows of each dataframe
map_df(nested_data$data[1:3], head, n = 2)
#> # A tibble: 6 x 23
#> binomial family genus species terrestrial marine freshwater aerial
#> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Abditom… Murid… Abdi… latide… 1 0 0 0
#> 2 Abeomel… Murid… Abeo… sevia 1 0 0 0
#> 3 Acerodo… Ptero… Acer… celebe… 0 0 0 1
#> 4 Acerodo… Ptero… Acer… humilis 0 0 0 1
#> 5 Acinony… Felid… Acin… jubatus 1 0 0 0
#> 6 Ailurop… Ursid… Ailu… melano… 1 0 0 0
#> # … with 15 more variables: life_habit_method <chr>, life_habit_source <chr>,
#> # mass_g <dbl>, mass_method <chr>, mass_source <chr>, mass_comparison <chr>,
#> # mass_comparison_source <chr>, island_endemicity <chr>, iucn_status <chr>,
#> # added_iucn_status <chr>, diet_plant <dbl>, diet_vertebrate <dbl>,
#> # diet_invertebrate <dbl>, diet_method <chr>, diet_source <chr>map accepts arguments to the function being mapped, such as in the example above, where head() accepts the argument n = 2.
map_dfr behaves the same as map_df.
# the same as above but with a pipe
nested_data$data[1:5] %>%
map_dfr(head, n = 2)
#> # A tibble: 10 x 23
#> binomial family genus species terrestrial marine freshwater aerial
#> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Abditom… Murid… Abdi… latide… 1 0 0 0
#> 2 Abeomel… Murid… Abeo… sevia 1 0 0 0
#> 3 Acerodo… Ptero… Acer… celebe… 0 0 0 1
#> 4 Acerodo… Ptero… Acer… humilis 0 0 0 1
#> 5 Acinony… Felid… Acin… jubatus 1 0 0 0
#> 6 Ailurop… Ursid… Ailu… melano… 1 0 0 0
#> # … with 4 more rows, and 15 more variables: life_habit_method <chr>,
#> # life_habit_source <chr>, mass_g <dbl>, mass_method <chr>,
#> # mass_source <chr>, mass_comparison <chr>, mass_comparison_source <chr>,
#> # island_endemicity <chr>, iucn_status <chr>, added_iucn_status <chr>,
#> # diet_plant <dbl>, diet_vertebrate <dbl>, diet_invertebrate <dbl>,
#> # diet_method <chr>, diet_source <chr>map_dfc binds the resulting 3 data frames of two rows each by column, and automatically repairs the column names, adding a suffix to each duplicate.
4.2.4 Working with list columns using map
The various map versions integrate well with list columns to make synthetic/summary data. In the example, the dplyr::mutate function is used to add three columns to the nested tibble: the number of rows, the mean mileage, and the name of the first car.
In each of these cases, the vectors added are generated using purrr functions.
# get the number of rows per dataframe, the mean mileage, and the first car
nested_data = nested_data %>%
mutate(
# use the int return to get the number of rows
n_rows = map_int(data, nrow),
# double return for mean mileage
mean_mass = map_dbl(data, function(df) {mean(df$mass_g)}),
# character return to get the heaviest member
first_animal = map_chr(data, function(df) {
arrange(df, -mass_g) %>%
.$binomial %>%
first()}
)
)
# examine the output
nested_data
#> # A tibble: 29 x 5
#> # Groups: order [29]
#> order data n_rows mean_mass first_animal
#> <chr> <list> <int> <dbl> <chr>
#> 1 Rodentia <tibble [2,306 × 23]> 2306 486. Neochoerus_aesopi
#> 2 Chiroptera <tibble [1,162 × 23]> 1162 49.1 Acerodon_jubatus
#> 3 Carnivora <tibble [313 × 23]> 313 47905. Mirounga_leonina
#> 4 Pilosa <tibble [34 × 23]> 34 785958. Megatherium_americanum
#> 5 Diprotodontia <tibble [183 × 23]> 183 40202. Diprotodon_optatum
#> 6 Cetartiodactyla <tibble [392 × 23]> 392 1854811. Balaenoptera_musculus
#> # … with 23 more rows4.2.5 Selective mapping using map variants
map_at and map_if work like other *_at and *_if functions. Here, map_if is used to run a linear model only on those tibbles which have sufficient data. The predicate is specified by .p.
In this example, the nested tibble is given a new column using dplyr::mutate, where the data to be added is a mixed list.
# split data by order number and run an lm only if there are more than 100 rows
nested_data = nest(data, data = -order)
nested_data = mutate(nested_data,
model = map_if(.x = data,
# this is the predicate
# which elements should be operated on?
.p = function(x){
nrow(x) > 100
},
# this is the function to use
# if the predicate is satisfied
.f = function(x){
lm(mass_g ~ diet_plant, data = x)
}))
# check the data structure
nested_data %>% head()
#> # A tibble: 6 x 3
#> order data model
#> <chr> <list> <list>
#> 1 Rodentia <tibble [2,306 × 23]> <lm>
#> 2 Chiroptera <tibble [1,162 × 23]> <lm>
#> 3 Carnivora <tibble [313 × 23]> <lm>
#> 4 Pilosa <tibble [34 × 23]> <tibble [34 × 23]>
#> 5 Diprotodontia <tibble [183 × 23]> <lm>
#> 6 Cetartiodactyla <tibble [392 × 23]> <lm>Some elements of the column model are tibbles, which have not been operated on because they have fewer than 100 rows (species). The remaining elements are lm objects.
4.3 More map variants
map also has variants along the axis of how many elements are operated upon. map2 operates on two vectors or list-like elements, and returns a single list as output, while pmap operates on a list of list-like elements.
The output has as many elements as the input lists, which must be of the same length.
4.3.1 Mapping over two inputs with map2
map2 has the same variants as map, allowing for different return types.
Here map2_int returns an integer vector.
# consider 2 vectors and replicate the simple vector addition using map2
map2_int(.x = 1:5,
.y = 6:10,
.f = sum)
#> [1] 7 9 11 13 15map2 doesn’t have _at and _if variants.
One use case for map2 is to deal with both a list element and its index, as shown in the example. This may be necessary when the list index is removed in a split or nest. This can also be done with imap, where the index is referred to as .y.
# make a named list for this example
this_list = list(a = "first letter",
b = "second letter")
# a not particularly useful example
map2(this_list, names(this_list),
function(x, y) {
glue::glue('{x} : {y}')
})
#> $a
#> first letter : a
#>
#> $b
#> second letter : b
# imap can also do this
imap(this_list,
function(x, .y){
glue::glue('{x} : {.y}')
})
#> $a
#> first letter : a
#>
#> $b
#> second letter : b4.3.2 Mapping over multiple inputs with pmap
pmap instead operates on a list of multiple list-like objects, and also comes with the same return type variants as map. The example shows both aspects of pmap using pmap_chr.
# operate on three different lists
list_01 = as.list(1:3)
list_02 = as.list(letters[1:3])
list_03 = as.list(rainbow(3))
# print a few statements
pmap_chr(list(list_01, list_02, list_03),
function(l1, l2, l3){
glue::glue('number {l1}, letter {l2}, colour {l3}')
})
#> [1] "number 1, letter a, colour #FF0000FF"
#> [2] "number 2, letter b, colour #00FF00FF"
#> [3] "number 3, letter c, colour #0000FFFF"4.4 Combining map variants and tidyverse functions
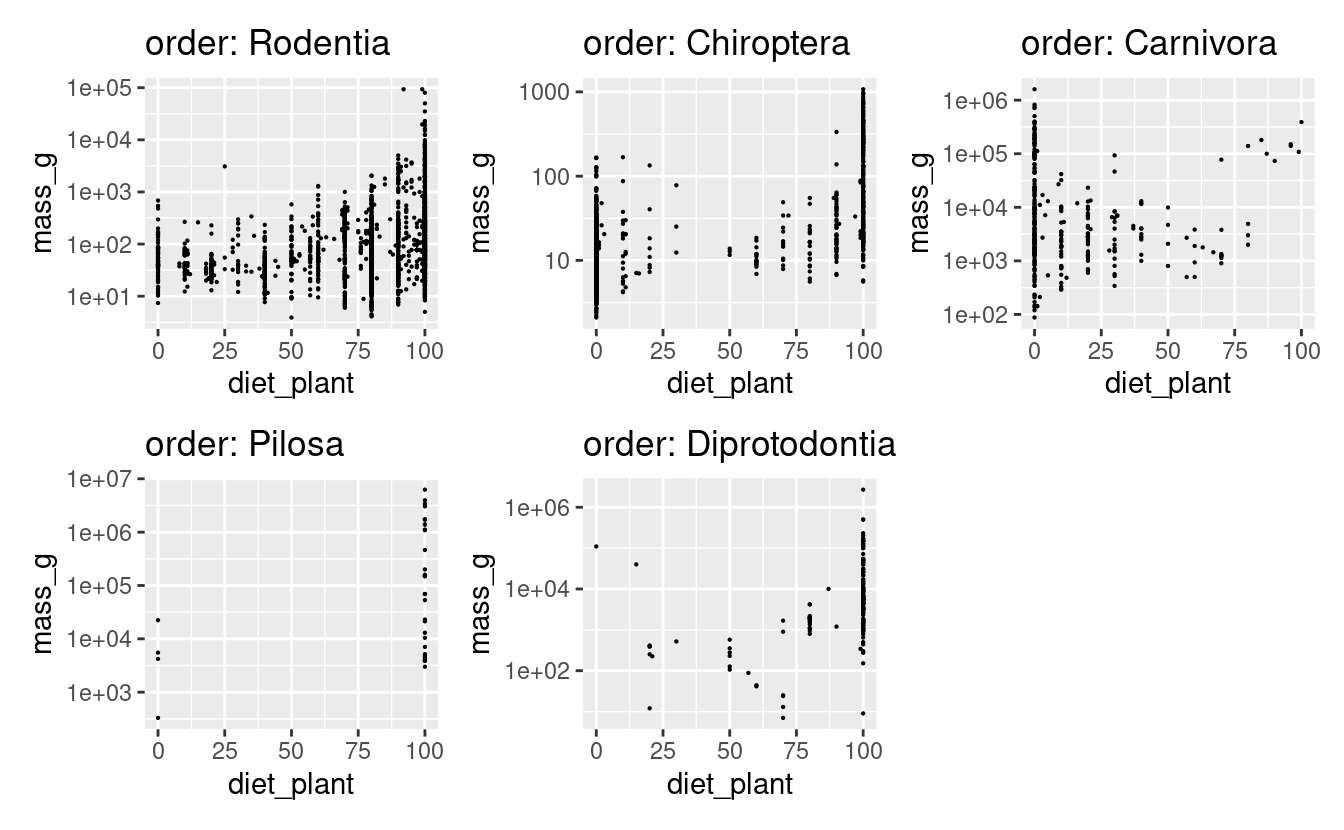
The example below shows a relatively complex data manipulation pipeline. Such pipelines must either be thought through carefully in advance, or checked for required output on small subsets of data, so as not to consume excessive system resources.
In the pipeline:
- The tibble becomes a nested dataframe by order (using
tidyr::nest), - If there are enough data points (> 100), a linear model of mass ~ plant diet is fit (using
purrr::map_if, andstats::lm), - The model coefficients are extracted if the model was fit (using
purrr::map&dplyr::case_when), - The model coefficients are converted to data for plotting (using
purrr::map,tibble::tibble, &tidyr::pivot_wider), - The raw data is plotted along with the model fit, taking the title from the nested data frame (using
purrr::pmap&ggplot2::ggplot).
nested_data <-
data %>%
tidyr::nest(data = -order) %>%
mutate(data,
model = map_if(.x = data,
# this is the predicate
# which elements should be operated on?
.p = function(x){
nrow(x) > 100
},
# this is the function to use
# if the predicate is satisfied
.f = function(x){
lm(mass_g ~ diet_plant, data = x)
})) %>%
mutate(m_coef = map(model,
# use case when to get model coefficients
function(x) {
dplyr::case_when(
"lm" %in% class(x) ~ {
list(coef(x))
},
TRUE ~ {
list(c(NA,NA))
}
)}),
# work on the two element double vector of coefficients
m_coef = map(m_coef, function(x){
tibble(coef = unlist(x),
param = c("intercept", "diet_plant")) %>%
pivot_wider(names_from = "param",
values_from = "coef")
}),
# work on the raw data and the coefficients
plot = pmap(list(order, data, m_coef), function(ord, x, y){
# pay no attention to the ggplot for now
ggplot2::ggplot()+
geom_point(data = x,
aes(diet_plant, mass_g),
size = 0.1)+
scale_y_log10()+
labs(title = glue::glue('order: {ord}'))
})
)4.5 A return to map variants
Lists are often nested, that is, a list element may itself be a list. It is possible to map a function over elements as a specific depth.
In the example, phylacine is split by order, and then by IUCN status, creating a two-level list, with the second layer operated on.
# use map to make a 2 level list
this_list = split(data, data$order) %>%
map(function(df){ split(df, df$iucn_status) })
# map over the second level to count the number of
# species in each order in each IUCN class
# display only the first element
map_depth(this_list[1], 2, nrow)
#> $Afrosoricida
#> $Afrosoricida$CR
#> [1] 1
#>
#> $Afrosoricida$DD
#> [1] 4
#>
#> $Afrosoricida$EN
#> [1] 7
#>
#> $Afrosoricida$EP
#> [1] 2
#>
#> $Afrosoricida$LC
#> [1] 32
#>
#> $Afrosoricida$NT
#> [1] 3
#>
#> $Afrosoricida$VU
#> [1] 84.5.1 Iteration without a return
map and its variants have a return type, which is either a list or a vector.
However, it is often necessary to iterate a function over a list-like object for that function’s side effects, such as printing a message to screen, plotting a series of figures, or saving to file.
walk is the function for this task. It has only the variants walk2, iwalk, and pwalk, whose logic is similar to map2, imap, and pmap. In the example, the function applied to each list element is intended to print a message.
this_list = split(data, data$order)
iwalk(this_list,
function(df, .y){
print(glue::glue('{nrow(df)} species in order {.y}'))
})
#> 57 species in order Afrosoricida
#> 313 species in order Carnivora
#> 392 species in order Cetartiodactyla
#> 1162 species in order Chiroptera
#> 39 species in order Cingulata
#> 74 species in order Dasyuromorphia
#> 2 species in order Dermoptera
#> 97 species in order Didelphimorphia
#> 183 species in order Diprotodontia
#> 465 species in order Eulipotyphla
#> 5 species in order Hyracoidea
#> 94 species in order Lagomorpha
#> 3 species in order Litopterna
#> 19 species in order Macroscelidea
#> 1 species in order Microbiotheria
#> 7 species in order Monotremata
#> 2 species in order Notoryctemorphia
#> 3 species in order Notoungulata
#> 7 species in order Paucituberculata
#> 24 species in order Peramelemorphia
#> 29 species in order Perissodactyla
#> 9 species in order Pholidota
#> 34 species in order Pilosa
#> 460 species in order Primates
#> 18 species in order Proboscidea
#> 2306 species in order Rodentia
#> 20 species in order Scandentia
#> 5 species in order Sirenia
#> 1 species in order Tubulidentata4.5.2 Modify rather than map
When the return type is expected to be the same as the input type, that is, a list returning a list, or a character vector returning the same, modify can help with keeping strictly to those expectations.
In the example, simply adding 2 to each vector element produces an error, because the output is a numeric, or double. modify helps ensure some type safety in this way.
vec = as.integer(1:10)
tryCatch(
expr = {
# this is what we want you to look at
modify(vec, function(x) { (x + 2) })
},
# do not pay attention to this
error = function(e){
print(toString(e))
}
)
#> [1] "Error: Can't coerce element 1 from a double to a integer\n"Converting the output to an integer, which was the original input type, serves as a solution.
A note on invoke
invoke used to be a wrapper around do.call, and can still be found with its family of functions in purrr. It is however retired in favour of functionality already present in map and rlang::exec, the latter of which will be covered in another session.
4.6 Other functions for working with lists
purrr has a number of functions to work with lists, especially lists that are not nested list-columns in a tibble.
4.6.1 Filtering lists
Lists can be filtered on any predicate using keep, while the special case compact is applied when the empty elements of a list are to be filtered out. discard is the opposite of keep, and keeps only elements not satisfying a condition. Again, the predicate is specified by .p.
# a list containing numbers
this_list = list(a = 1, b = -1, c = 2, d = NULL, e = NA)
# remove the empty element
# this must be done before using keep on the list
this_list = compact(this_list)# use discard to remove the NA
this_list = discard(this_list, .p =is.na)
# keep list elements which are positive
keep(this_list, .p = function(x){ x > 0 })
#> $a
#> [1] 1
#>
#> $c
#> [1] 2head_while is bit of an odd case, which returns all elements of a list-like object in sequence until the first one fails to satisfy a predicate, specified by .p.
4.6.2 Summarising lists
The purrr functions every, some, has_element, detect, detect_index, and vec_depth help determine whether a list passes a certain logical test or not. These are seldom used and are not discussed here.
4.6.3 Reduction and accumulation
reduce helps combine elements along a list using a specific function. Consider the example below where list elements are concatenated into a single vector.
This can also be applied to data frames. Consider some random samples of mtcars, each with only 5 cars removed. The objective is to find the cars present in all 10 samples.
The way reduce works in the example below is to take the first element and find its intersection with the second, and to take the result and find its intersection with the third and so on.
# sample mtcars
mtcars = as_tibble(mtcars, rownames = "car")
sampled_data = map(1:10, function(x){
sample_n(mtcars, nrow(mtcars)-5)
})
# get cars which appear in all samples
sampled_data = reduce(sampled_data,
dplyr::inner_join)accumulate works very similarly, except it retains the intermediate products. The first element is retained as is. accumulate2 and reduce2 work on two lists, following the same logic as map2 etc.
Both functions can be used in much more complex ways than demonstrated here.
4.6.4 Miscellaneous operation
purrr offers a few more functions to work with lists (or list like objects). prepend works very similarly to append, except it adds to the head of a list. splice adds multiple objects together in a list. splice will break the existing list structure of input lists.
flatten has a similar behaviour, and converts a list of vectors or list of lists to a single list-like object. flatten_* options allow the output type to be specified.
this_list = list(a = rep("a", 3),
b = rep("b", 4))
this_list
#> $a
#> [1] "a" "a" "a"
#>
#> $b
#> [1] "b" "b" "b" "b"
# use flatten chr to get a character vector
flatten_chr(this_list)
#> [1] "a" "a" "a" "b" "b" "b" "b"transpose shifts the index order in multi-level lists. This is seen in the example, where the iucn_status goes from being the index of the second level to the index of the first.
4.7 Lists of ggplots with patchwork
The patchwork library helps compose ggplots, which will be properly introduced in the next session. patchwork usually works on lists of ggplots, which can come from a standalone list, or from a list column in a nested dataframe.
The example below shows the latter, with the data data frame from earlier.